R,
Code,
Analytics,
Apache Spark,
Data Science,
h2o,
Predictive Analytics,
Tutorials,
Sparkling Water,
Machine Learning
Table of Contents
Need Help? Get in Touch!
At Red Oak Strategic, we utilize a number of machine learning, AI and predictive analytics libraries, but one of our favorites is h2o. Not only is it open-source, powerful and scalable, but there is a great community of fellow h2o users that have helped over the years, not to mention the staff leadership at the company is very responsive (shoutout Erin LeDell and Arno Candel).
However, as we were building models on large datasets, as we regularly do, our data science team began running into scale issues while running these models and scores locally, so we needed to move to a more distributed solution. h2o’s Sparkling Water, leveraging the h2o algorithms on top of Apache Spark, was a perfect solution. As an AWS Partner, we wanted to utilize the Amazon Web Services EMR solution, but as we built these solutions, we also wanted to write up a full tutorial end-to-end for our tasks, so the other h2o users in the community can benefit. Shoutout as well to Rahul Pathak at AWS for his help with EMR over the years.
This blog will cover the following topics, allowing you to start from no AWS infrastructure, building to a powerful, scaled and distributed predictive analytics and machine learning setup, built on top of Apache Spark:
- Create an AWS EMR cluster (we assume you already have an AWS account)
- SSH and install h2o Sparkling Water and necessary dependencies
- Make some configuration tweaks
- Connect from your local machine in R/RStudio
- Model and score!
- Spin down your cluster
I will note: we started with h2o’s built-in instructions for spinning up an EMR cluster, but made a few tweaks to our own setup, and to upgrade versions.
Create an AWS EMR Cluster
There are two methods to creating your EMR cluster: through the AWS CLI (command line tool), or via the Amazon Web Services UI. We prefer to use the CLI for flexibility, so we will utilize that method. If you’d like to use the UI, these instructions outline those steps. Again, I am assuming your AWS CLI is configured and setup, but if not, follow these instructions to do so.
Make note that below you will need to customize your KeyName , the subnets/security group information, tags , as well as the number of instances (InstanceCount )/size of your cluster (I’ve set mine at 6), as well as the InstanceType for your head node, as well as the workers.
Note: This assumes Spark 2.3.0, Sparkling Water 2.3.5, and h2o 3.18.10 are the versions you wish to install. To install other versions, change the commands as needed.
Things to review/update:
- tags
- ec2-attributes
- KeyName
- SecurityGroup / SubnetId
- release-label (which version of the basic software do you want?)
- Note: emr-5.13 has Spark 2.3.0 installed
- name
- instance-groups
- InstanceCount
- InstanceType
- region
- profile (set to profile name from your own ~/.aws/credentials file )
$ aws emr create-cluster \
--applications Name=Hadoop Name=SPARK Name=Zeppelin Name=Ganglia \
--no-visible-to-all-users \
--tags 'Owner=mark' 'Purpose=H2O Sparkling Water Deployment' 'Name=h2o-spark' \
--ec2-attributes '{"KeyName":"your_key_name_here","InstanceProfile":"EMR_EC2_DefaultRole","SubnetId":"subnet-xxxxxx","EmrManagedSlaveSecurityGroup":"sg-xxxxxx","EmrManagedMasterSecurityGroup":"sg-xxxxxxx","AdditionalMasterSecurityGroups":["sg-xxxxxx"]}' \
--service-role EMR_DefaultRole \
--release-label emr-5.13.0 \
--name 'h2o-spark' \
--instance-groups '[{"InstanceCount":6,"EbsConfiguration":{"EbsBlockDeviceConfigs":[{"VolumeSpecification":{"SizeInGB":100,"VolumeType":"gp2"},"VolumesPerInstance":1}]},"InstanceGroupType":"CORE","InstanceType":"c4.8xlarge"},{"InstanceCount":1,"EbsConfiguration":{"EbsBlockDeviceConfigs":[{"VolumeSpecification":{"SizeInGB":100,"VolumeType":"gp2"},"VolumesPerInstance":1}]},"InstanceGroupType":"MASTER","InstanceType":"c4.8xlarge"}]' \
--scale-down-behavior TERMINATE_AT_INSTANCE_HOUR \
--region us-east-2 \
--profile mark
After a few minutes, your cluster should be available and viewable, either through the EMR UI, or from the command line.
$ aws emr list-clusters
SSH and Configure Cluster
Something that we wanted to customize a bit was the shell script that runs after spinning up our cluster. This is something long term that we can automate as a bootstrap script, but for purposes of this tutorial we’re listing the full file.
First, SSH into the head node (from your terminal, ensure SSH port is open from your IP on the master node security group):
$ ssh -i your_key_name_here.pem hadoop@ec2.your.master.node.ip.here
Run the following on the head node after SSHing.
set -x -e
sudo python -m pip install --upgrade pip
sudo python -m pip install --upgrade colorama
sudo ln -sf /usr/local/bin/pip2.7 /usr/bin/pip
sudo pip install -U requests
sudo pip install -U tabulate
sudo pip install -U future
sudo pip install -U six
#Scikit Learn on the nodes
sudo pip install -U scikit-learn
version=2.3.5
h2oBuild=5
SparklingBranch=rel-${version}
mkdir -p /home/hadoop/h2o
cd /home/hadoop/h2o
echo -e "\n Installing sparkling water version $version build $h2oBuild "
wget https://h2o-release.s3.amazonaws.com/sparkling-water/${SparklingBranch}/${h2oBuild}/sparkling-water-${version}.${h2oBuild}.zip &
unzip -o sparkling-water-${version}.${h2oBuild}.zip 1> /dev/null &
export MASTER="yarn"
echo -e "\n Rename jar and Egg files"
mv /home/hadoop/h2o/sparkling-water-${version}.${h2oBuild}/assembly/build/libs/*.jar /home/h2o/sparkling-water-${version}.${h2oBuild}/assembly/build/libs/sparkling-water-assembly-all.jar
mv /home/hadoop/h2o/sparkling-water-${version}.${h2oBuild}/py/build/dist/*.egg /home/h2o/sparkling-water-${version}.${h2oBuild}/py/build/dist/pySparkling.egg
echo -e "\n Creating SPARKLING_HOME env ..."
export SPARKLING_HOME="/home/hadoop/h2o/sparkling-water-${version}.${h2oBuild}"
export PYTHON_EGG_CACHE="~/"
export SPARK_HOME="/usr/lib/spark"
Launch the Sparkling Water Cluster
Once your install script is finished successfully, navigate to the sparkling-water install directory:
$ cd sparkling-water-2.3.5
To launch the Spark cluster, type the following command with any edits for the specific EMR setup you initialized (eg. instances number, cores, memory):
$ bin/sparkling-shell \ --master yarn \ --conf spark.deploy-mode=cluster \ --conf spark.executor.instances=6 \ --conf spark.executor.cores=36 \ --conf spark.executor.memory=40g \ --conf spark.driver.memory=40g \ --conf spark.scheduler.maxRegisteredResourcesWaitingTime=1000000 \ --conf spark.ext.h2o.fail.on.unsupported.spark.param=false \ --conf spark.dynamicAllocation.enabled=false \ --conf spark.sql.autoBroadcastJoinThreshold=-1 \ --conf spark.locality.wait=30000 \ --conf spark.scheduler.minRegisteredResourcesRatio=1
Once you see the Spark/Scala prompt run these commands to start h2o:
import org.apache.spark.h2o._ val h2oContext = H2OContext.getOrCreate(spark) import h2oContext._
If you’ve configured everything correctly, your output will look like this (with different IP addresses for your instances – the head and the worker machines):
scala> import org.apache.spark.h2o._ import org.apache.spark.h2o._ scala> val h2oContext = H2OContext.getOrCreate(spark) h2oContext: org.apache.spark.h2o.H2OContext = Sparkling Water Context: * H2O name: sparkling-water-hadoop_application_1234567890_0001 * cluster size: 6 * list of used nodes: (executorId, host, port) ------------------------ (1,ip-172-00-00-11.us-east-2.compute.internal,54321) (3,ip-172-00-00-12.us-east-2.compute.internal,54321) (4,ip-172-00-00-13.us-east-2.compute.internal,54321) (6,ip-172-00-00-14.us-east-2.compute.internal,54321) (5,ip-172-00-00-15.us-east-2.compute.internal,54321) (2,ip-172-00-00-16.us-east-2.compute.internal,54321) ------------------------ Open H2O Flow in browser: http://ip-172-00-00-11.us-east-2.compute.internal:54321 (CMD + click in Mac OSX) scala> import h2oContext._ import h2oContext._
The H2O Flow will be available at the public IP of the head node at port 54321. For example:
your.ec2.instance.public.ip:54321
Something to check: make sure port 54321 is open in your security groups, so you can connect from your IP.
Connect from your local RStudio
To utilize our cluster, we now can connect from RStudio.
library(tidyverse) library(sparklyr) library(h2o) library(janitor) options(rsparkling.sparklingwater.version = "2.3.5") library(rsparkling) h2o.init(ip = "ip address of master node", port = 54321)
Now, instead of utilizing the local R cluster on your laptop or desktop, you can utilize the distributed version on AWS.
Get Data Into Your Cluster
Something we wanted to add here: the speed of transferring files locally to your R cluster can significantly impact performance. We would recommend utilizing the s3 ImportFile function, which significantly speeds up that data transfer.
airlinesURL <- "https://s3.amazonaws.com/h2o-airlines-unpacked/allyears2k.csv" airlines.hex <- h2o.importFile(path = airlinesURL, destination_frame = "airlines.hex")
Monitor Your Cluster
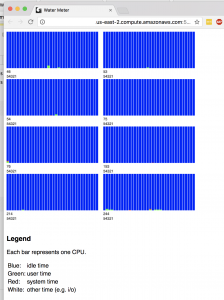
h2o includes a very nice cluster monitoring tool called “WaterMeter” which allows you to view and monitor the cluster CPU usage. In this screenshot, we have an 8-node cluster – look at all of those beautiful CPUs!
Terminate Your Cluster
Once you’ve finished your work, and you want to spin down or terminate your cluster, use the following command from your local machine (in your Terminal):
$ aws emr terminate-clusters --cluster-ids j-3KVXXXXXXX7UG
Wrapping Up
Again, we want to thank the h2o community as well as h2o and Amazon Web Services staff for their assistance and guidance. There are almost unlimited possibilities for this implementation, but we’d love to hear from you about ways you’re using Sparkling Water or h2o in your work. Or, if you have questions/comments for the Red Oak Strategic team, please leave them below and we’ll get back in touch. Thanks for reading.
Contact Red Oak Strategic
From cloud migrations to machine learning & AI - maximize your data and analytics capabilities with support from an AWS Advanced Tier consulting partner.
Related Posts
Machine Learning

Tyler Sanders
Machine Learning

Tyler Sanders
Ready to get started?
Kickstart your cloud and data transformation journey with a complimentary conversation with the Red Oak team.





